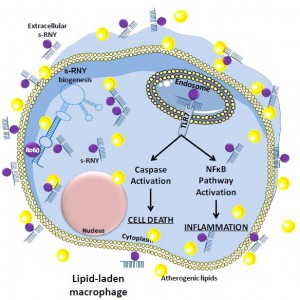
Investigation of novel mechanisms of miRNA-dependent gene expression control
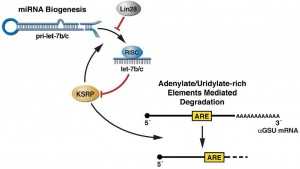
We recently demonstrated a direct role for let-7b/c to regulate KSRP expression, which in turn controls let-7 processing in competition with Lin28, and the turnover rate of the pituitary terminal differentiation marker aGSU leading to an upregulation of its expression. These findings defined a hierarchical mechanism by which during organogenesis the upregulation of specific miRNAs controls the expression of KSRP and the subsequence fine-tuning of the processing for selected miRNAs and decay rate of mRNAs encoding for differentiation markers.Currently, we are using proteomic and high-throughput sequencing approaches coupled with bioinformatics to study the protein/miRNA code to orchestrate the post- transcriptional programme(s) of regulated gene expression.